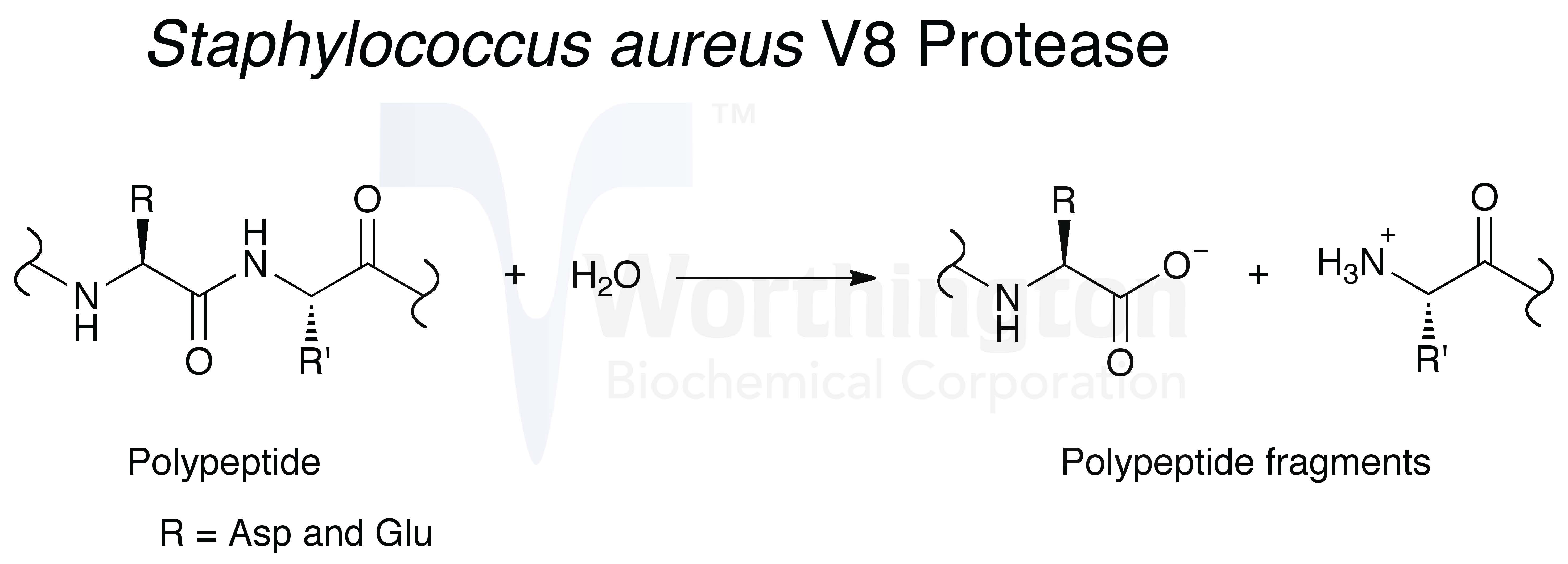
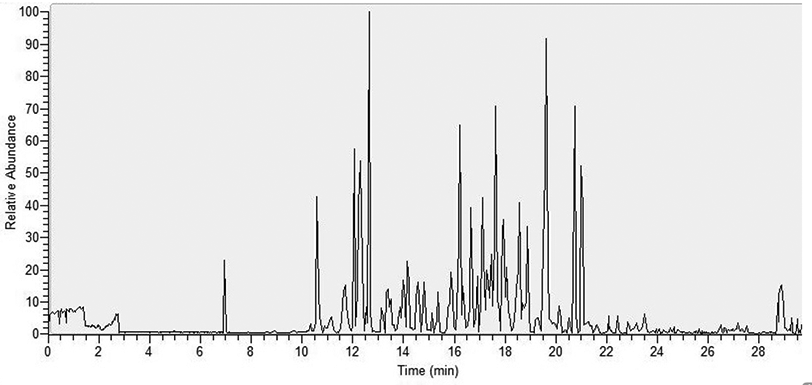
One-Step Isolation of Protein C-Terminal Peptides from V8 Protease-Digested Proteins by Metal Oxide-Based Ligand-Exchange Chromatography | Analytical Chemistry

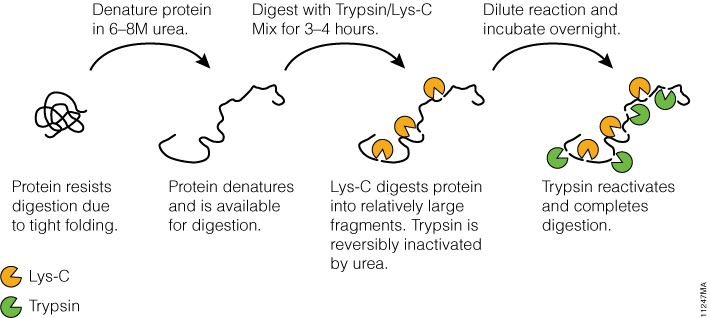
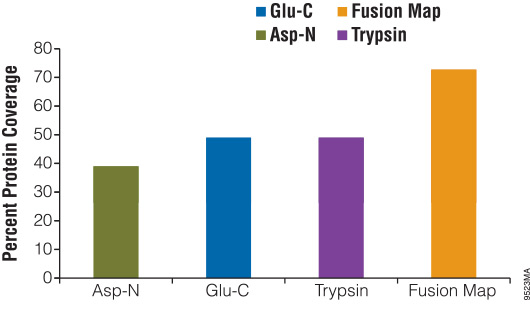
Proteomics Using Protease Alternatives to Trypsin Benefits from Sequential Digestion with Trypsin | Analytical Chemistry
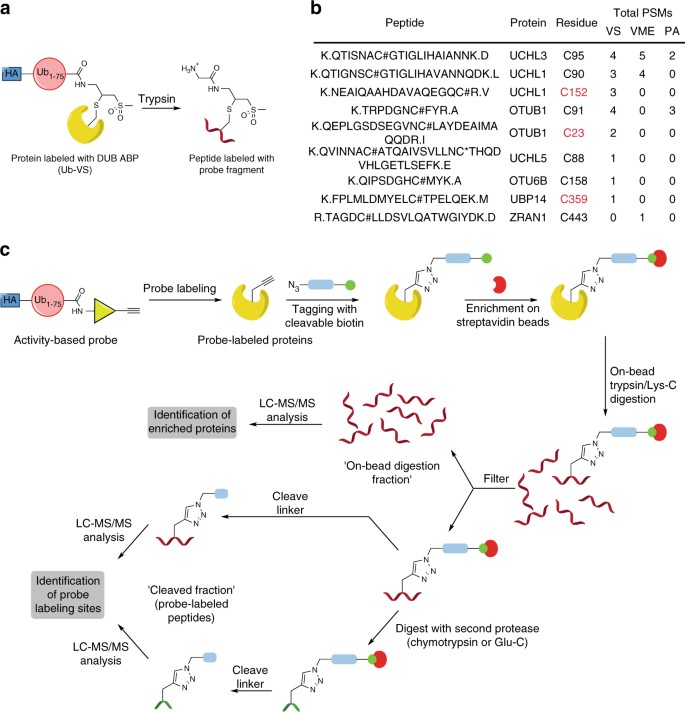
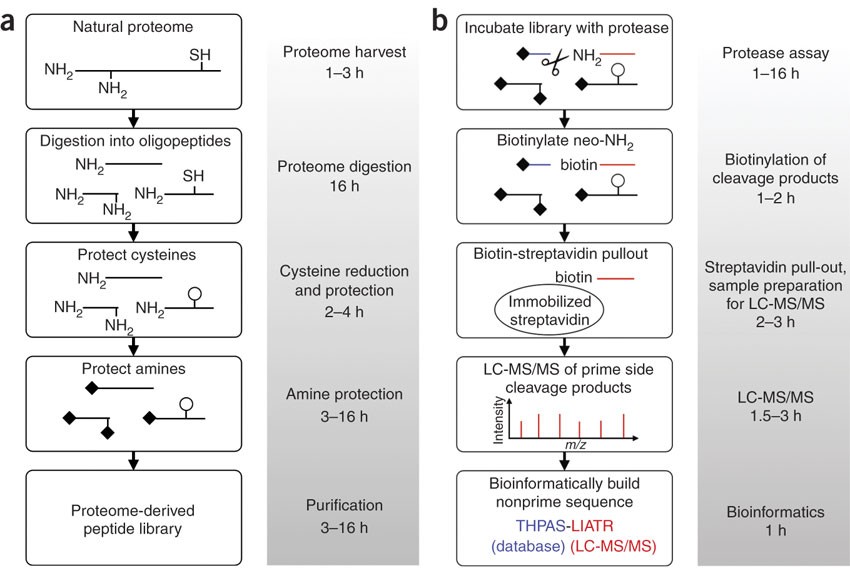
Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes | Nature Communications

Proteomics Using Protease Alternatives to Trypsin Benefits from Sequential Digestion with Trypsin | Analytical Chemistry
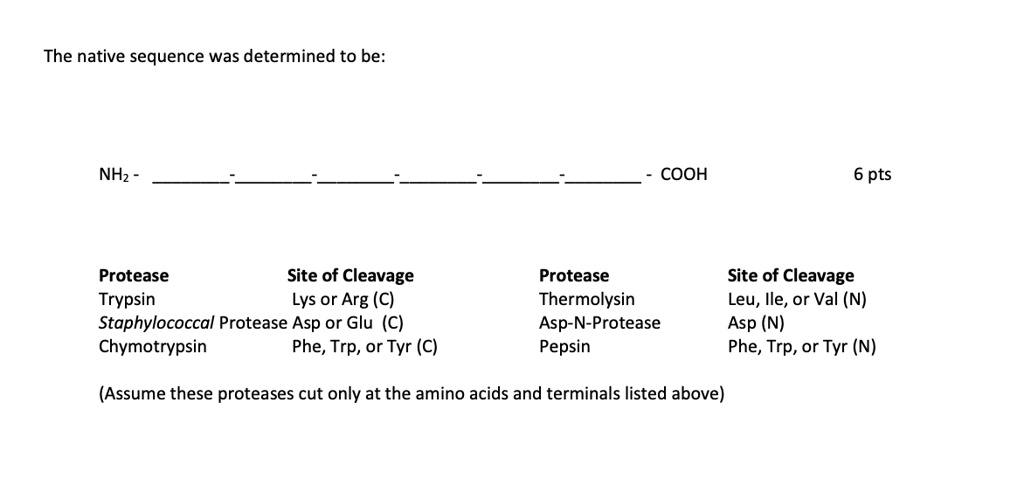
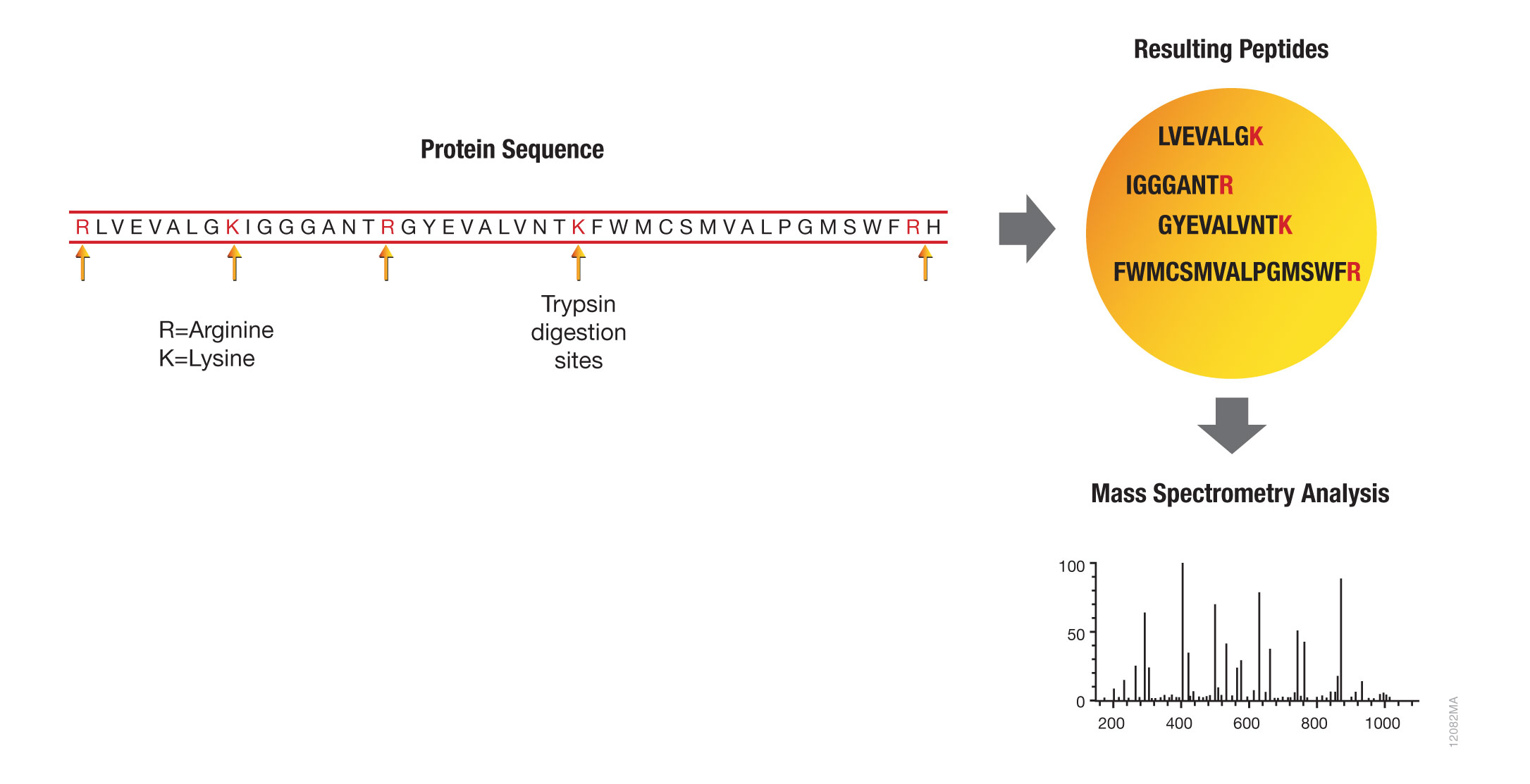
SOLVED: The native sequence was determined to be: COOH 6 pts Protease Site of Cleavage Trypsin Lys or Arg (C) Staphylococcal Protease Asp or Glu (C) Chymotrypsin Phe, Trp, or Tyr (C)

Substrate cleavage specificity of legumain, GluC, and trypsin. IceLogos... | Download Scientific Diagram